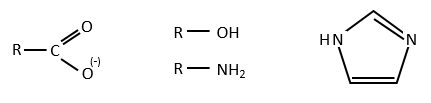
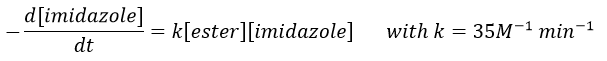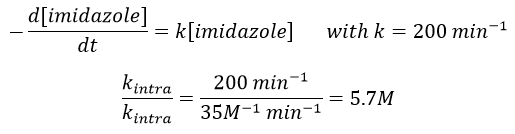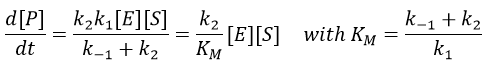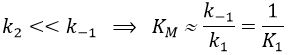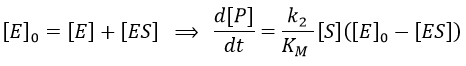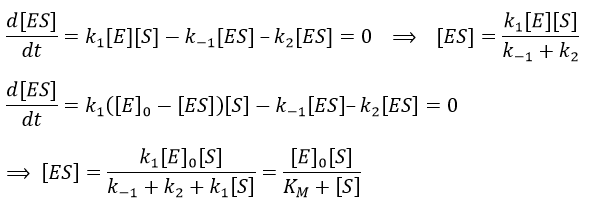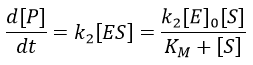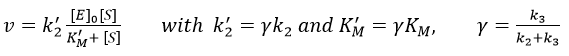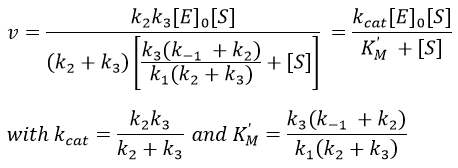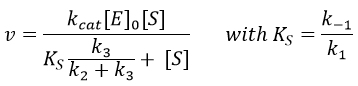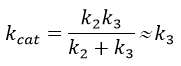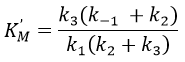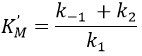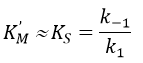It can be done several ways
- catalysis by proximity (entropic contribution)
- electrophile and nucleophile catalyses
- general basic catalysis
Catalysis by proximity
We compare here an intramolecular process with an intermolecular process.
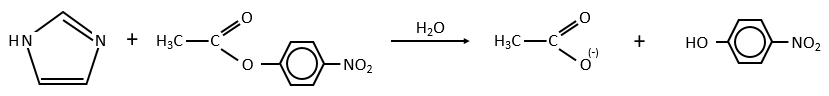
Intermolecular
Imidazole is incorporated into many important biological molecules. One of them is the histidine that is present in many proteins and enzymes. The presented reaction is of the second order.
The p-nitrophenol is in equilibrium with its deprotonated form at neutral pH, form that can be detected via spectrometry of absorption.
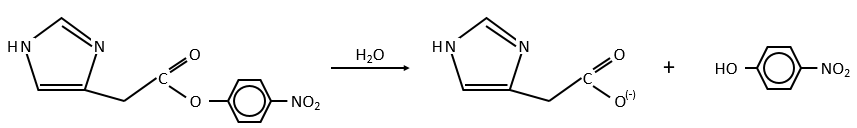
Intramolecular
The fact that the catalyst was initially bound to the reactant is thus equivalent to the presence of 5.7M of the ester. The fact that the catalyst is already well positioned plays a role and there is also an entropic factor: from one molecule we produce two molecules and there is no need to bind the reactant to the catalyst anymore.
Nucleophilic catalysis
There are nucleophilic groups on enzymes. They can thus act as nucleophile catalysts (see previously).
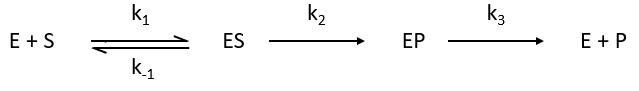
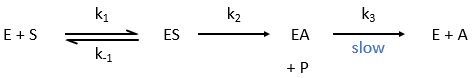
Equation of Michaelis-Menten
The enzymes are in small quantity in comparison to their substrates. The enzyme forms a complex with its substrate and then acts on it to form the product.
From the section on an equilibrium followed by an irreversible reaction, we know that
In this section, we have seen that there are two limit cases: the irreversible reaction is very fast or the equilibrium has time to be reached. In this case, the equilibrium has time to be established.
The quantity of enzyme is conserved over the time, in its lone form or complexed with the substrate.
We used the hypothesis of stationarity on [ES].
The formation of the product P is given by
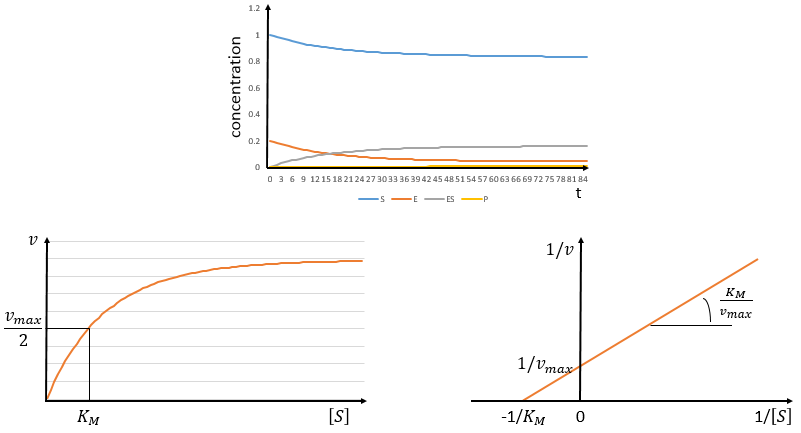
The speed of the reaction depends thus strongly on the concentration of the substrate up to a given point where [S]>>KM. The speed reaches then a plateau at v=k2[E]0. The maximum of the speed is thus reached when all the enzymes are active simultaneously ([ES]=[E]0).
The half of this speed is reached for a concentration [S]=KM.
By plotting 1/v vs 1/[S] (before the plateau), we find a straight line from which we can determine KM and k2.
If we consider one more step in the reaction, the formation of the product when it is still complexed with the enzyme, i.e. before the separation of the species, we will have a correction factor γ to add.
Case of the α-chymotrypsin
This enzyme is active in the digestive system where it hydrolyses polypeptides. It is active in this basic environment (pH between 8 and 9) and breaks the amide bonds between the peptides.
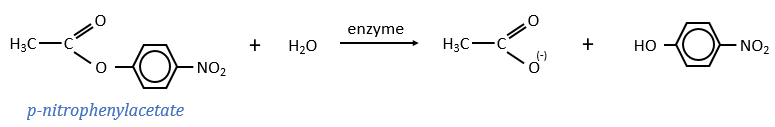
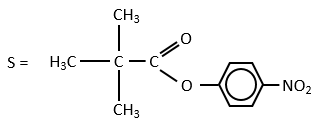
For the experiments, we substitute the polypeptides by an ester, the p-nitrophenylacetate.
The p-nitrophenol is detected due to its yellow colour (405nm) and the acetate is not detected. The observations of the experiment are particular: the enzyme acts very fast at the beginning of the experiment and then the speed reaches a constant level. It is thus the opposite of what we have seen just above. If we look at the small times, the production of phenol is directly proportional to the quantity of enzyme. It seems that it is in this step of the reaction that we have vmax=k2[E]0. There should be a step after the release of the phenol that inhibits the enzyme. This step is the release of the acetate which is much slower than the release of the phenol.
where P=p-nitrophenol, A=phenylacetate and k3<<k2.
As a result, when the phenol is released, the enzyme is not yet available to cleave another substrate. It has first to release the acetate, which is much slower. At the beginning of the reaction, we observe an important and quick increase of [phenol] that is quickly released by the enzyme. Before this moment, all the enzymes can do their job, what explains the large speed of reaction. After, one fraction of the enzymes is still occupied by one fragment of the substrate and cannot react until the acetate is released. The speed of the reaction is thus limited by the third step of the reaction.
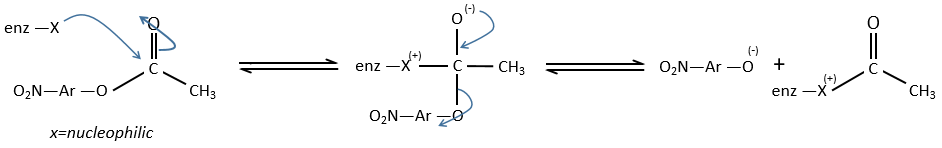
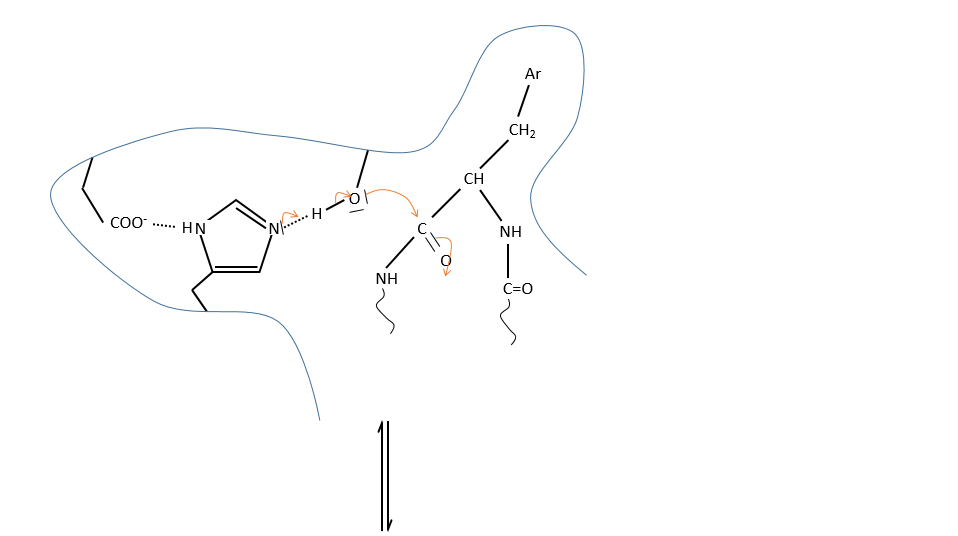
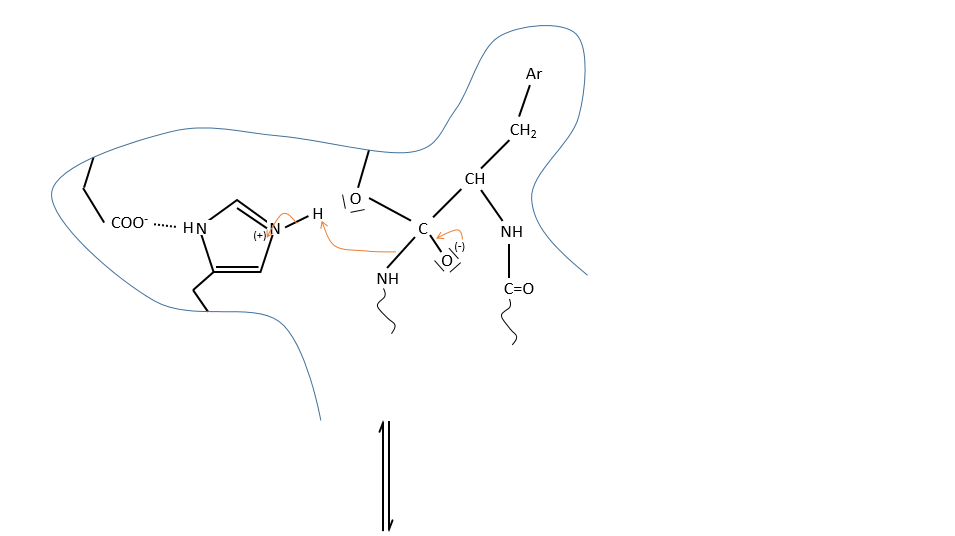
We can assume that there is a nucleophilic site on the enzyme where the substrate binds at the level of the carbonyl.
We can show that the intermediate acylenzyme is correct with the use of a more hindered substrate.
The steric hindrance and the inductive character of the methyl’s slow down the reaction, and thus they increase the life time of the acyled intermediate (~200 minutes). We can isolate the intermediate as crystals and analyse them.
We can use a better nucleophile than water to remove the enzyme.
This acetate (acylhydroxamate) can chelate with Fe3+ to form a red species that can be detected.
The speed of the reaction in four steps is
If k-1>>k2, then
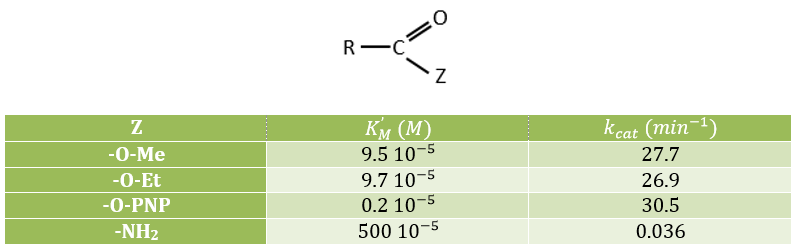
Comparison of several substrates
We compare here the speeds of reaction for several esters and for one amide, the natural substrate of the enzyme.
The three esters have the same speed of reaction because k3<<k2. As a result, kcat limits the speed of the reaction.
The leaving group has no real influence on the speed of the reaction. Indeed, OPNP is a better leaving group (K’M~KS k3/k2à k2=2500k3, in comparison to k2=50k3 for OEt or OMe) but kcat do not change by much. In the case of the amide, i.e. the normal substrate of the enzyme, k2<<k3 and the speed of the reaction depends on the leaving group. There is no accumulation of E.A.
As k2<<k3,
We can also assume that k2<<k-1 because the equilibrium is established. Consequently,
KS is the constant of enzyme-substrate dissociation. This constant is approximatively identical no matter the substrate.
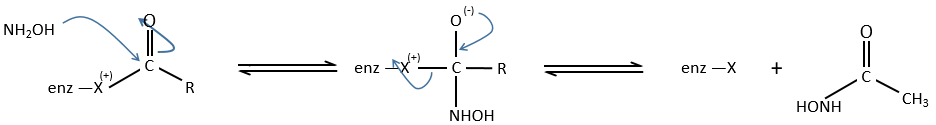
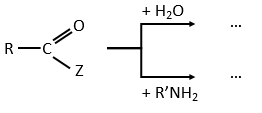
An additional verification can be done through a competition between nucleophiles. If we compare the kinetics of reaction using water and an amine RNH2. The two species are in competition to react with the substrate.
The amine is a better nucleophile but the concentrations play a role in the competition. We observe that in the case of esters, the speed of reaction increases with the ratio amine/water. The speed increases as long as kW+kN<k2. When kW+kN>k2, the acylation becomes the limiting step of the reaction. In the case of amides, the competition doesn’t change anything: E.A doesn’t accumulate so there is no competition.
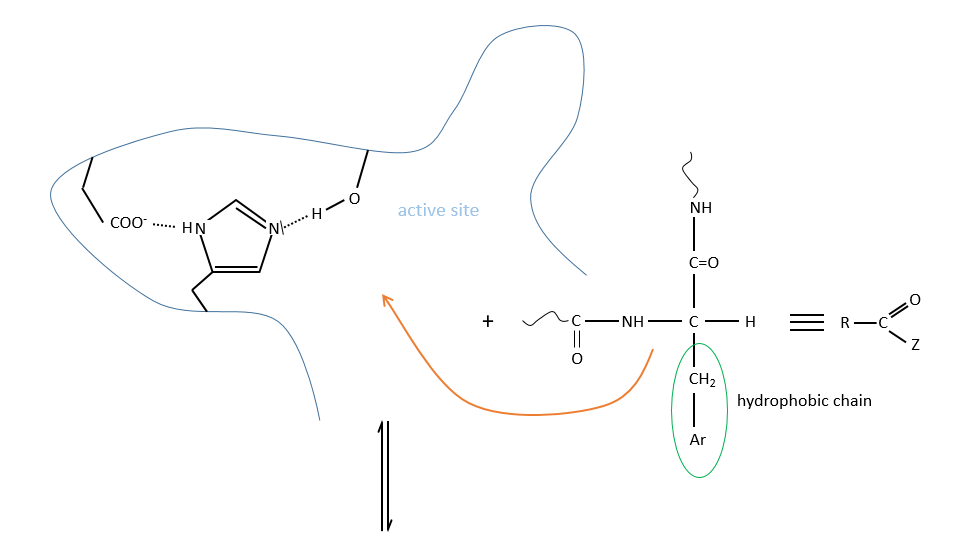
Structure and activity of the chymotrypsin
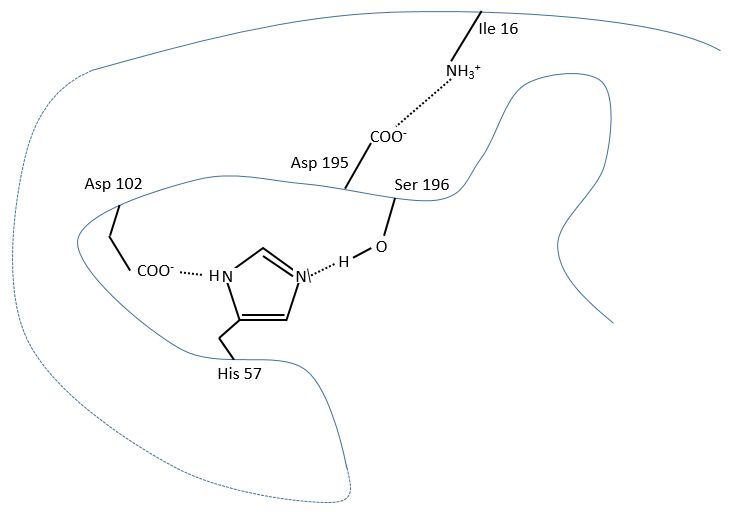
It is possible to determine the amino acids of the active site of proteases by X rays. There are 3 of them:
The enzyme is active between pH 7 and 9 so the active AA of the chymotrypsin is the histidine 57 but it is not the nucleophile in the previous reactions. It has been shown by the introduction of a marked inhibitor in substitution of the substrate to be able to observe the intermediate by looking at the peptide bound to the inhibitor after the hydrolysis of all peptide bonds.
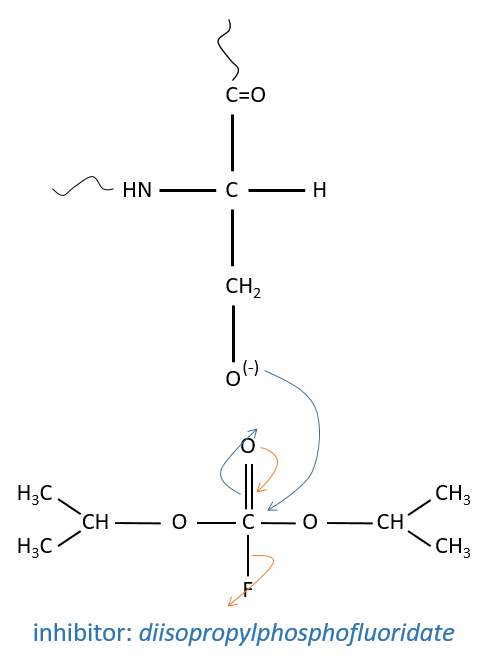
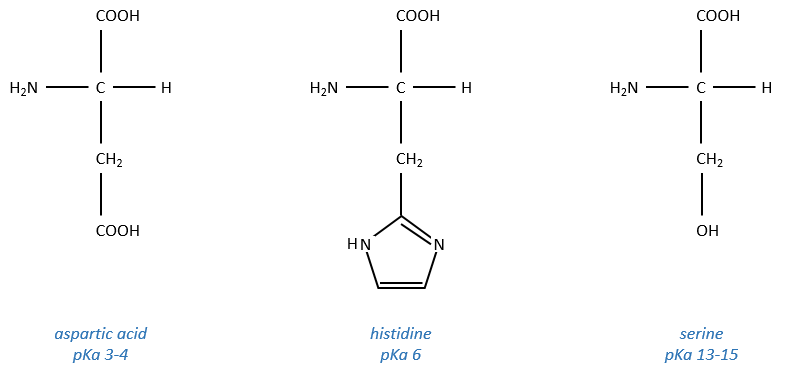
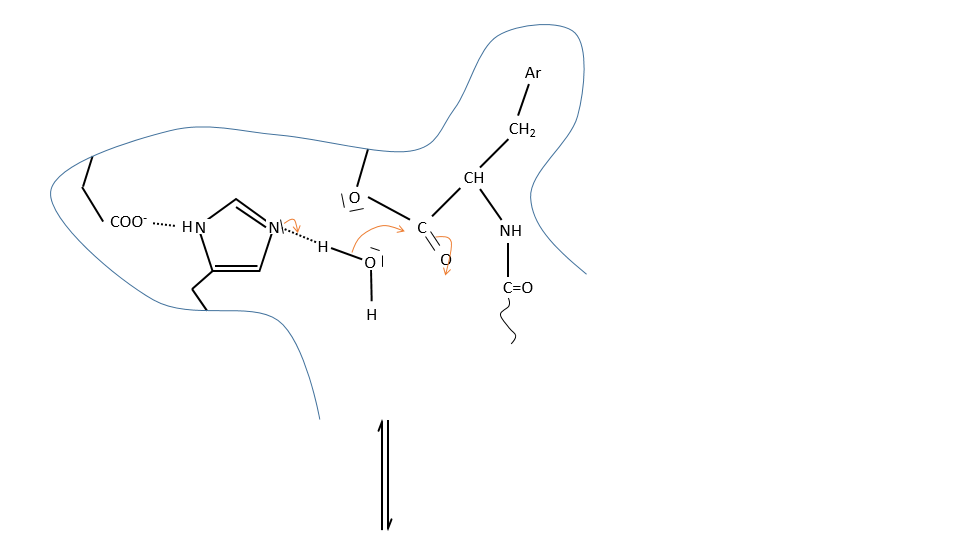
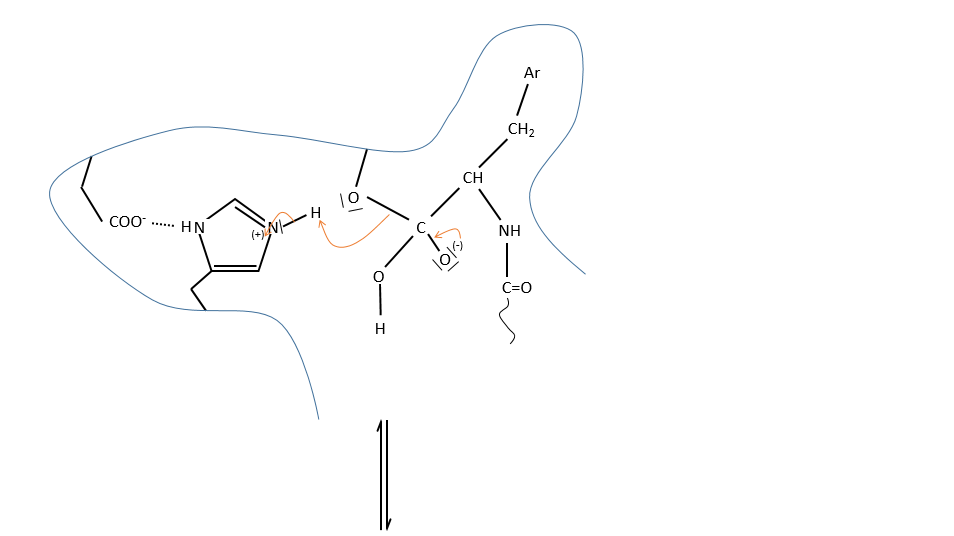
It is the serine that binds the inhibitor but it has a pKa between 13-15. So why is it active? Something increased the nucleophilic character of the serine. Histidine is active in the range of pH in which the enzyme works and it boosts the activity of the serine through general basic catalysis.
The enzyme is active at this pH because there is a change of conformation. The active site is not available in other conformations.
In more acid conditions, H3O+ gets inside the pocket containing the active site and blocks it. In more basic conditions, the conformation change because the NH3+ of the Ile 16 is not protonated anymore. As a result it doesn’t interact with the COO– of the Asp 195 and the conformation of the active site changes to deny its access to the substrate.
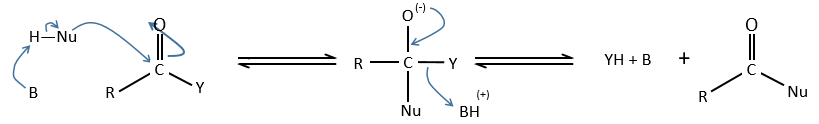
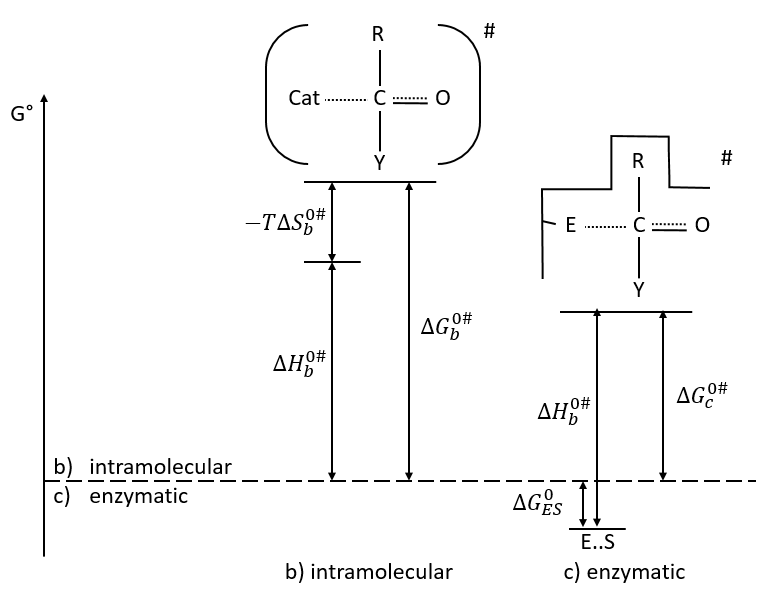
Comparison between the enzymatic, inter- and intramolecular general basic catalyses
a. intermolecular general basic catalysis
The first step of the reaction is the limiting one. The catalysis helps the reaction to take place with an important decrease of the enthalpy of reaction. As a result, ∆G0 decreases too but not by that much because of the entropic term: 3 species are involved in the reaction, what requires some organisation and there is thus a decrease of entropy.
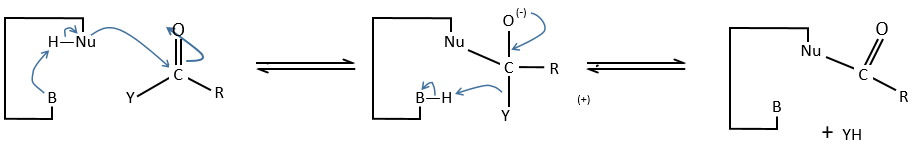
b. intramolecular general basic catalysis
The reaction is faster than for the intermolecular catalysis because the entropic term is smaller.
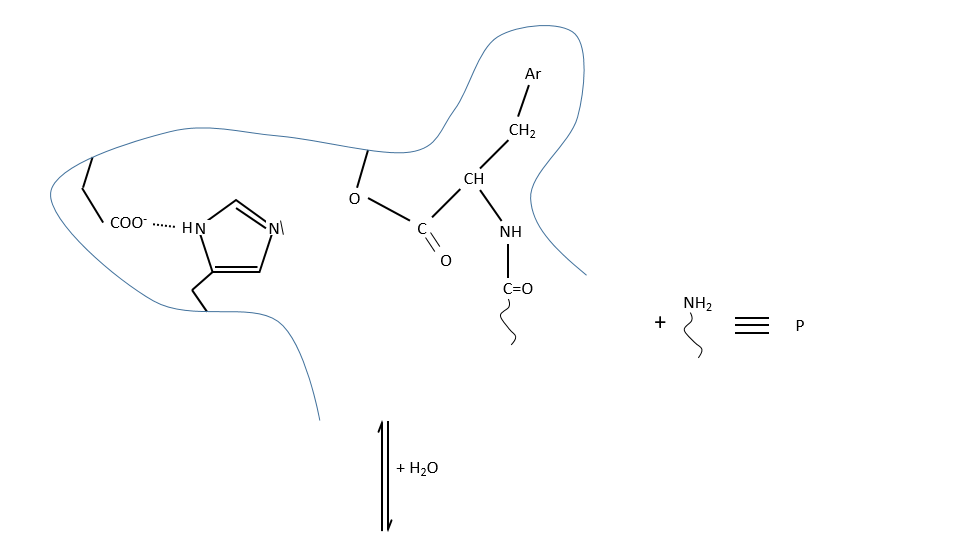
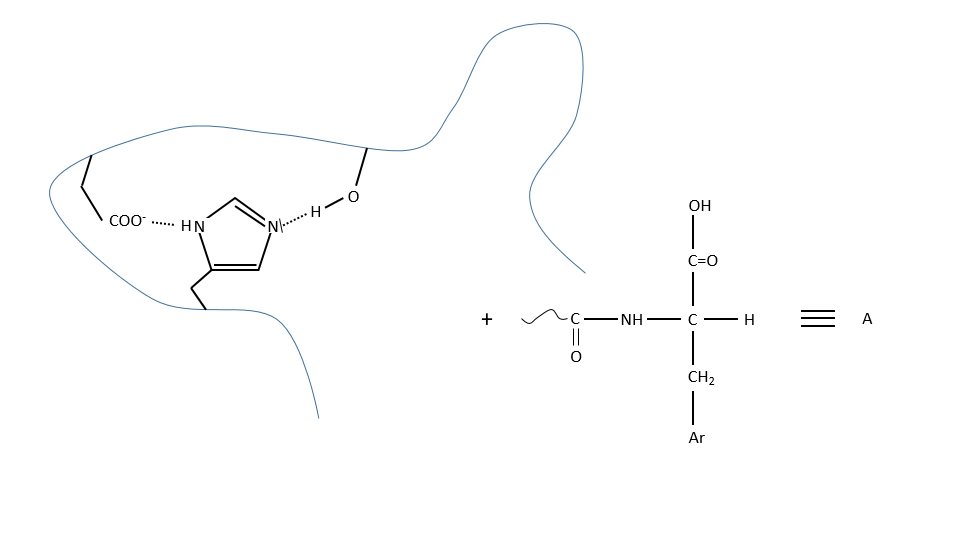
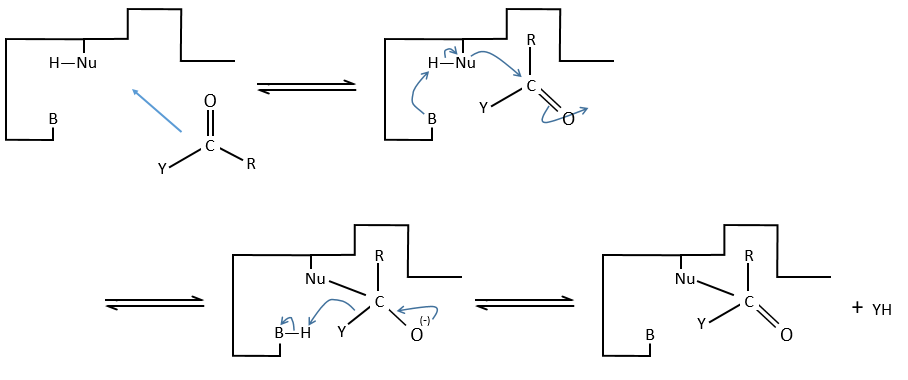
c. enzymatic catalysis
The product can be removed from the enzyme with the addition of H2O to restore the catalyst. There is an additional step in this catalysis: the fixation of the substrate in the active site.
Let’s focus on this first approach step: the entropy term is positive but it is smaller than the enthalpic term (negative).
In comparison with the intramolecular catalysis, we start thus from a lower G° and the enthalpy of formation of the intermediate species are identical (or approximatively equal).
The entropic term of the intermediate b is approximatively equivalent to the one of the ES intermediate.
As ∆H°ES<0, there is a decrease of ∆G° in the process because of the favourable positioning of the substrate in the active site. ∆H°ES can be large with regard to ∆Gb°#, so the difference of speed between the two processes can be large. For the case of the α- chymotrypsin, the enzymatic hydrolysis is 108 times faster at pH=8.