1) Definition
The DNA replication is one of the key mechanisms of cell cycle. It is a highly regulated biological process in which new synthesis and repair mechanisms ensure the integrity of the genome.
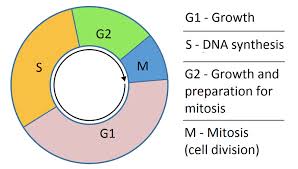
S phase or replication phase follows the G1 phase during which the cell synthesizes all the elements necessary for this replication. The DNA duplication takes about 8 hours.
2) substrates ot the replication
2.1. template strand
The double helix of DNA is separated into two DNA strands. Each strand is then used as template for the synthesis of a new strand.
Replication produces 2 hybrid molecules formed from a parent strand and a daugther strand : replication is semi-conservative.
2.2. Nucleotides
- Nucleotides: dATP, dCTP, dTTP dGTP providing both:
the substrate: the nucleoside monophosphate,
Energy for linking nucleotides between them.
2.3. Enzymes
a) there are several DNA polymerases according to their replicative activity:
– alpha: Start Replication
– delta : replication of the nuclear genome
– Gamma: replication of the mitochondrial genome
– beta: DNA repair
– epsilon: replication of telomeres
b) other proteins are also required during replication
3) Mechanism of replication
3.1. origines of replication
In eukaryotes, replication is initiated at many replication origins called OriC, unlike prokaryotes in which there is a single origin of replication.
Eukaryotic cells must replicate their entire genome for each cell cycle. To carry out this process in a short time, the initiation of replication is made at several OriC or “replication origins” along the chromosome (about 10000) in independent units of replication called replicon (30-300 bp).
In eukaryotes, the best characterized replication origins are those of the yeast. They are named for Autonomously Replicating Sequence ARS. These ARS regions rich in AT, possess fixation sites for the ORC complex.
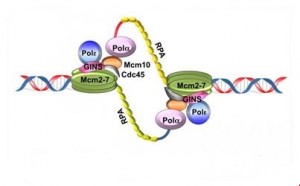
Then other proteins capable of unwinding the DNA double helix are recruited (by activation of helicases).
3.2. Activation of replication origins
3.2.1. pre-replication complex formation
Replication origins are recognized by protein complexes “ORC” (origin recognizing Complex ) formed of six subunits, each of which is fixed on each replication origin.
Following this fixation, two other factors, Cdc6 / Cdc18 and CDT1, join the first complex to recruit six protein complexes (MCM2, 3,5,6,7,8 )
3.2.2.Activation of the pre-replication complex
1) Activation of CDK
2) phosphorylation / activation of proteins including helicases (Mcm2-7)
3) helicases unwind the DNA double helix and separate the two strands.
The unwinding of the DNA and separating of the two strands exposes single-stranded DNA regions, which are stabilized with protein RPA (replication protein A) and accessible to enzymes and proteins required for replication.
4) Recruitment of DNA polymerases and other proteins
3.3. DNA polymerases
DNA polymerases (or deoxynucleotidyl transferase) are the enzymes responsible for the polymerization of the nucleotides in DNA replication. Prokaryotic DNA polymerases are 3 types (I, II and III) and eukaryotic DNA polymerases of 5 types (α, β, δ, ε and γ).
3.3.1 DNA polymerase α
The DNA polymerase α (α- primase) synthesizes primers which is a short RNA segment (10 base pairs). These short RNA fragments are then elongated by a DNA fragment (20 to 40 nucleotides long). An RNA-DNA primer is created.
3.3.2. DNA polymerase δ
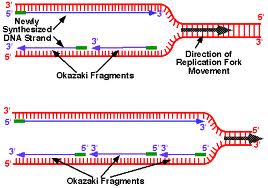
The synthesis of DNA is always in the 5 ‘to 3′, so this requires the presence of a leading strand (primary) which is the strand read in the fork direction and a lagging strand (secondary) is the strand read in the opposite direction of the fork and is said discontinuous strand. There is talk of semi-discontinuous replication.
A complex RF-C / PCNA (replication factor C/ Proliferating cell nuclear antigen) is fixed on the 3’OH end of the primer RNA / DNA and dissociates DNA polymerase α of the DNA template, leaving instead the DNA polymerase δ which recognizes the complex RFC / PCNA and be responsible for the synthesis of continuous strand.
On the strand with discontinuous synthesis, DNA polymerase α-primase which was previously dissociated, restarts the synthesis of a new primer RNA / DNA. On this DNA strand, the enzyme synthesizes multiple small DNA fragments in the prologation of primers, called “Okazaki fragments” (along approximately 1000 to 5000 nucleotides).
As the progression of the replication goes on :
helicase unwind the DNA double helix and topo- isomerase reduces twisting of the DNA upstream of the forks.
3.3.3. DNA ligase
It reconstructs the phosphoester bond between the 3′-OH carbon and phosphate-5′ of two adjacent nucleotides in a DNA strand.
It is also involved in many DNA repair processes.
3.4 The topoisomerases
Type I
– Performs a cut on one of the strands of DNA.
– Transition
– Release the twist.
– No ATP necessary.
Type I I
– Performs a cut on the two strands of DNA.
– Requires ATP.