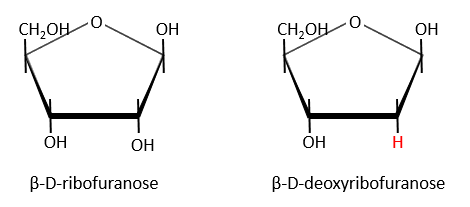
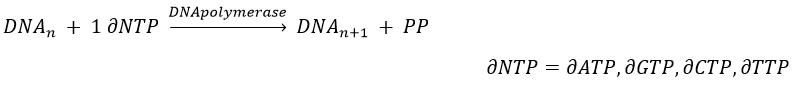Nucleic acids are composed of monosaccharides connected by phosphoester liaisons and wearing a base. One monosaccharide and its base are called one nucleoside. One monosaccharide, its base and one phosphate are called a nucleotide. RNA and DNA are composed of one similar monosaccharide, the ribofuranose, with the difference that the 2’ has an H on the DNA in place of an OH for the RNA. It is the reason of their names: the ribonucleic acid (RNA) and the deoxyribonucleic acid (DNA).
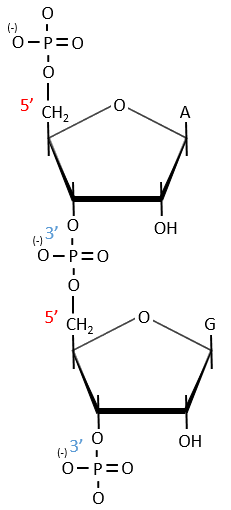
Monosaccharides are bound together in 3’-5’ and the base is bound by the OH on 1’. There is a total of 5 bases.
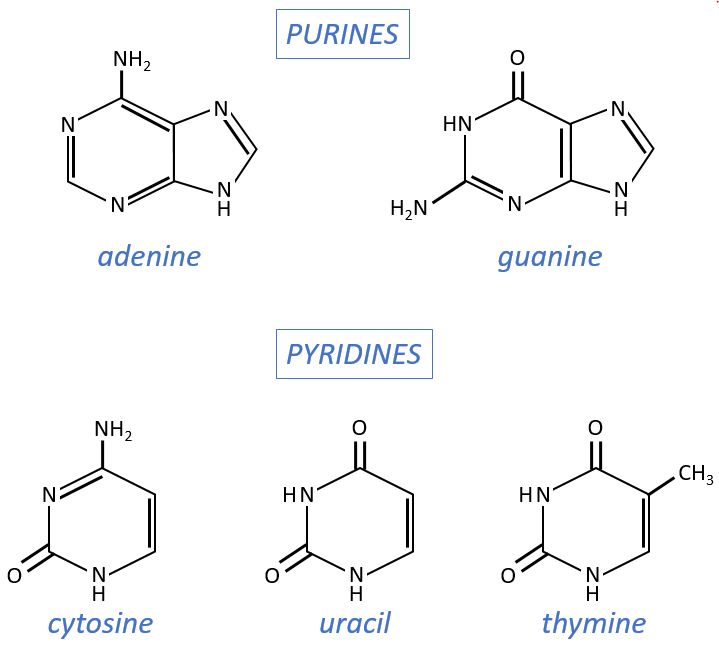
- purine bases: they are composed of two aromatic cycles with nitrogen atoms in the cycles. They absorb the UV at 260nm.
- adenine (A)
- guanine (G)
- pyrimidine bases: they are composed of one aromatic cycle and also absorb UV.
- uracil (U, only found in RNA)
- thymine (T, only found in DNA)
- cytosine (C)
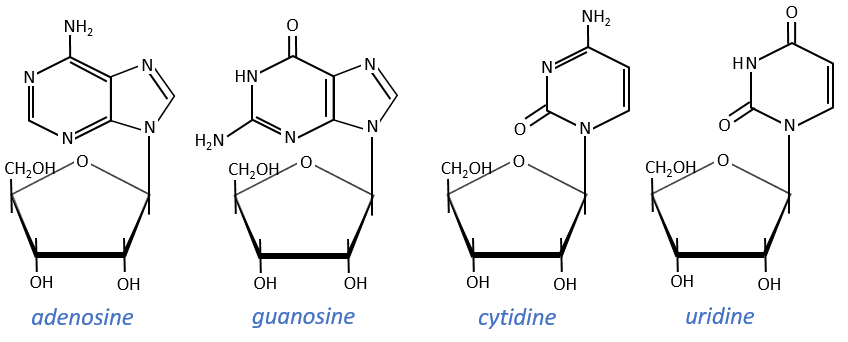
Nucleosides are the combination of one base with one monosaccharide. The names of nucleoside are directly related to their base.
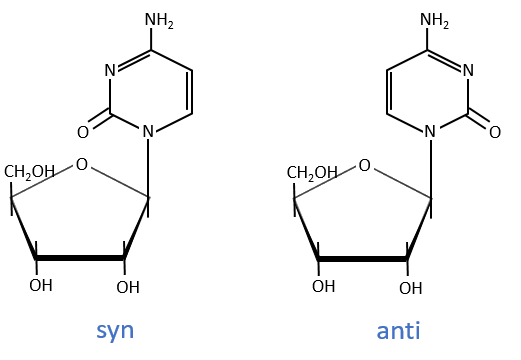
The same is true for the deoxynucleosides. The prefix deoxy is just added to the name of the corresponding nucleoside. There is a stereochemistry in the orientation of the base and the monosaccharide. The liaison can either be in anti or in syn.
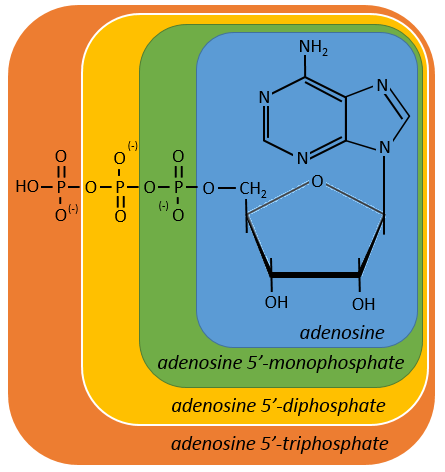
Nucleotides are the combination of one nucleoside with one or more phosphates (in chains). The phosphates can be placed on 5’ or on 3’ by an ester liaison and phosphates are bound as acid anhydride of acid. The liaison between two nucleosides is a phosphodiester liaison.
One example: two important nucleotides are the adenosine diphosphate (ADP) and the adenosine triphosphate (ATP).
If the base was a deoxy~, we write a ∂ (or d) before the name (ex: ∂ATP)
Nucleotides absorb in the UV the same way nucleosides do.
A chain of nucleic acids has a polarity. It goes from the 5’ towards the 3’. A chain is written in this order in abbreviated:
The chains are also build following this order.
Cleavage of the chain
- By total acid hydrolysis, using a 12M HCl solution. It cleaves all the liaisons between the monosaccharides, the bases and the phosphates.
It is the harsh method, the following methods are gentler.
- By depurination: we work at pH=1.6 and at 37°C. The cleavage only works for the DNA and only break β-N-glycosidic bonds. β-N-glycosidic bonds are hydrolytically cleaved releasing a nucleic base, adenine or guanine, respectively.
- hydrolysis for RNA: we use NAOH 0.1M at 25°C (during 15hours). It only cleaves the RNA chains because the reaction involves an intermediate species that needs the OH in 3’
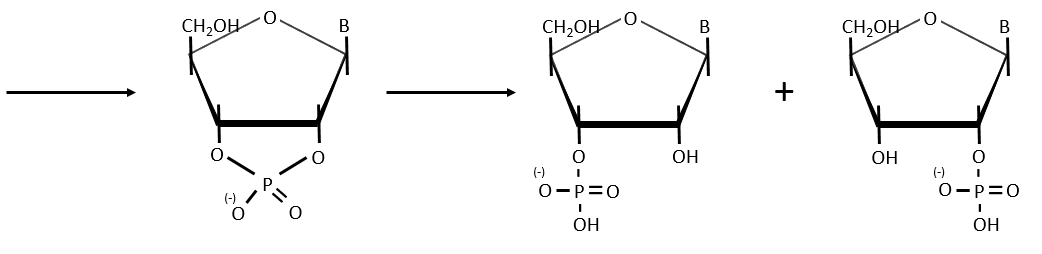
- enzymatic cleavages: some enzymes are specialised in the cleavage of nucleic chains. There are of two kinds: endonucleases and exonucleases. The firsts cleave liaisons inside the chain and the seconds cleave the chains starting at one extremity. It can go from 3’ to 5’ or from 5’ to 3’, depending on the enzyme. The extremity has to be free of phosphate to be cleaved. For instance, the phosphodiesterase that we have in the spleen only works if the 5’ extremity is ended by OH. It will free nucleosides 3’-phosphate. The phosphodiesterase that we find in the venom of snakes cleaves the 3’ extremity and forms nucleosides 5’-phosphate.
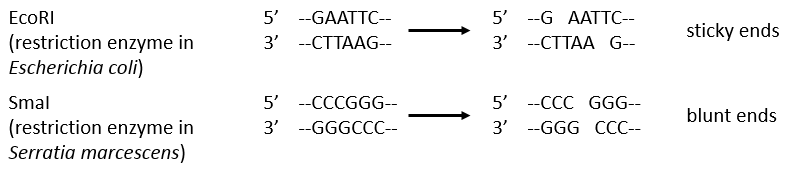
Endonucleases are usually not specific and give nucleosides 3’ phosphates. Yet there are some specific endonucleases that recognise a particular pattern in the chain. For instance, endonucleases of restriction are enzymes that we find in bacteria’s to protect themselves from bacteriophages. Bacteriophages are viruses that attack bacteria’s and try to inject their own DNA inside the DNA chain of the bacteria. The enzyme recognise a palindrome in the sequence of the DNA. It is called a site of restriction. This site is cleaved in a special way.
The resulting extremities may be sticky or blunt, depending if the cleavage is done at the same base or not on the two strands. The term sticky was chosen because they are easily joined back together by a ligase if there are several unpaired nucleotides.
The DNA of the bacteria is protected from this cleavage because its DNA was methylated by a methyltransferase after/during its formation.
Structure of the DNA
DNA has a helical structure that was determined by several experiments.
- Chayaff: he was studying the composition of the DNA in term of bases. He performed a total acid hydrolysis and analysed then the proportions of A, C, T and G. The result of its experiment was that the proportion of A is equal to the one of T and that the proportion of G was equal to the proportion of C. Moreover, the sum A+G=C+T.
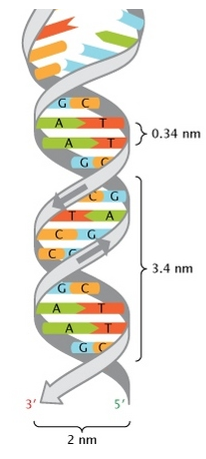
- Wilkin and Franklin: They performed the XR diffraction of the DNA. They observed particular pattern with a two characteristic lengths of 0.34nm and of 3.4nm.
- Watson and Crick: From the experiment of Wilkin and Franklin, they gave a model of structure for the DNA: a double helix.
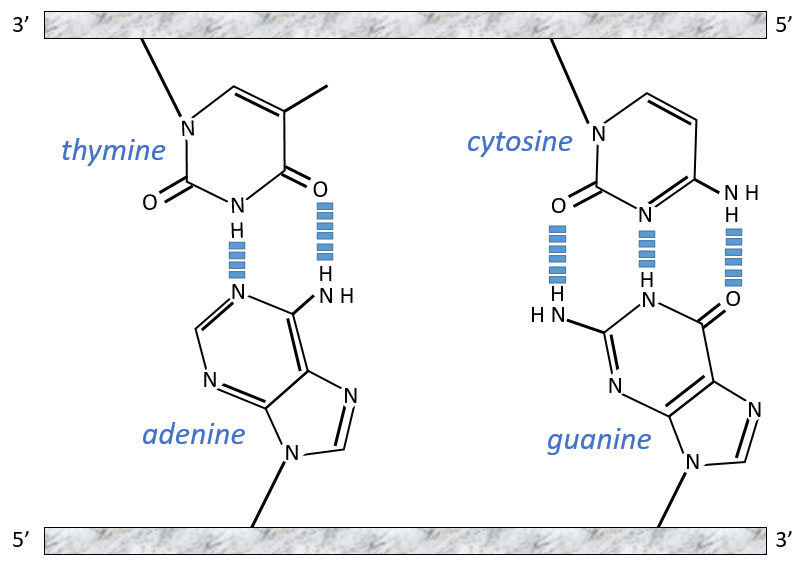
The chains of monosaccharides are at the outside of the double helix and the bases are facing each other at the inside of the helix. Each base can only be pairing one other base: A with T and C with G. It explains the equivalences in proportion.
The helix is stable thanks to the H bonds between the bases. There are 2 H bonds between A and T and 3 between C and T.
As they are surrounded by the hydrophobic chain of monosaccharides, the bases are protected from water that could interact with the H bonds linking the bases. There are also electrostatic interaction between the bases.
A heating (80-90°C) can separate the two “strands” of DNA from each other. When separated, they absorb more light than when they are coupled and we can observe a hyperchromic effect at 260nm.
The sites where enzymes cleave the DNA are always full of A or/and T. It is easier to open the helix here because there are less H bonds between the bases.
The length of the DNA varies between species and is not in helix for the prokaryotes for which the DNA is circular. The longest genome belongs to the “triton”
Replication of the DNA
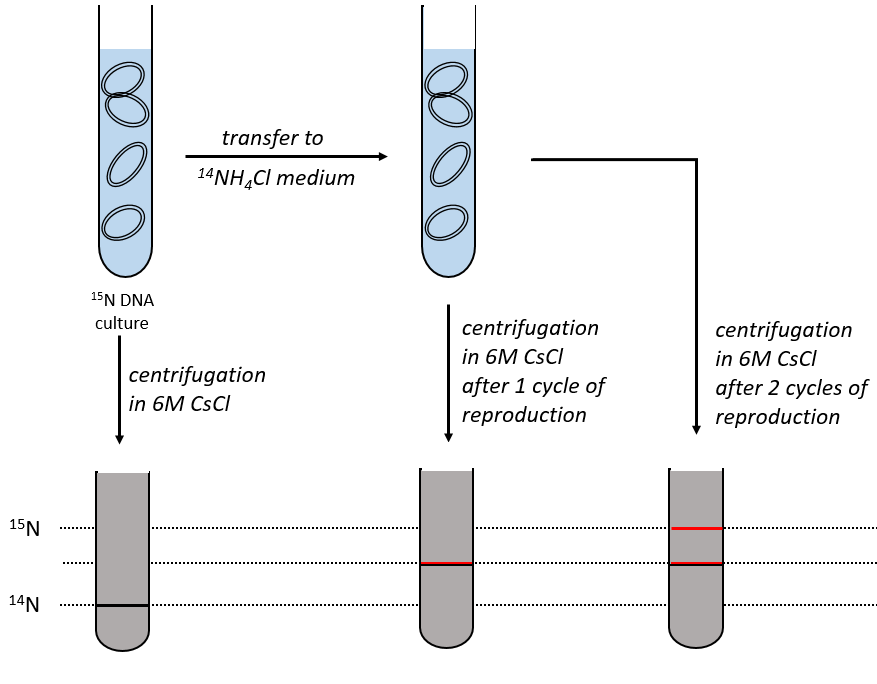
During the mitosis, the cell splits in two cells, the DNA is reproduced and each new cell possesses the same genome than the cell they are made of, the mother cell. The DNA can spontaneously reform itself if it was denatured as it was shown by the experiment of Miseber and Slahl: they placed some bacteria’s in a solution the composition of which is controlled: the single source of nitrogen in the solution is NH4Cl and the nitrogen were marked isotopes. The bacteria’s form the bases of the DNA with this nitrogen. After a while, the DNA is centrifuged with a dense salt of CsCl. During the centrifugation, the salt goes to the bottom of the tube and the DNA stops at its density. As there are two isotopes of nitrogen (14N and 15N), there will be two layers of DNA with different densities.
We can thus isolate the light DNA from the heavy DNA. If we place them respectively in a solution of culture with 14N and with 15N, they can replicate. From the solutions, we extract some DNA, put them together and denature them (T=100°C). The strands of DNA are thus separated. When the solution is cooled down, the strands pair together but not necessarily with the strand they were paired with previously. After centrifugation, there are still the layers of DNA that were obtained previously but there is now a third layer between the two other, corresponding to the binding of one 14N strand with one 15N strand.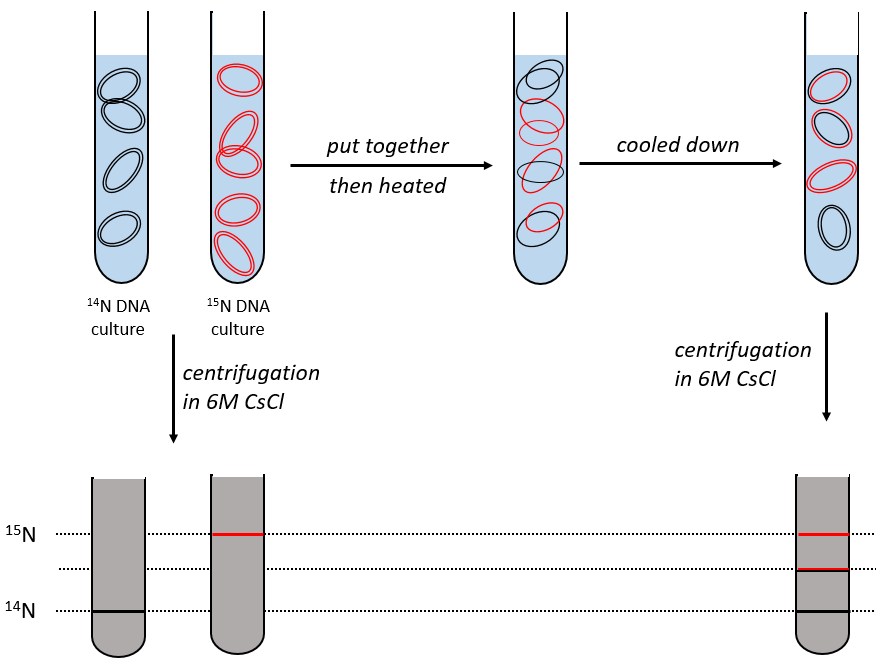
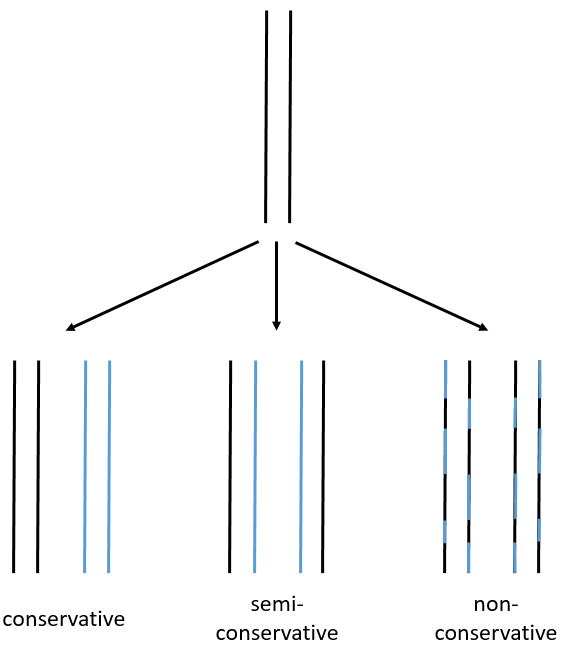
We can imagine three ways of replication for the DNA: we consider a mother strand of DNA and one daughter strand that is the replication of the mother strand.
- conservative replication: the two strands of the mother DNA remain paired after the replication.
- semi-conservative replication: one strand of the mother DNA pairs with one strand of daughter DNA.
- non-conservative replication: the sequences of mother and daughter DNA are mixed in both strands.
The replication of the DNA is semi-conservative. It can be confirmed by placing the 15DNA in a culture with 14N. If the replication was conservative, we would only obtain two layers of different densities. If the replication was non-conservative, there would be a continuous layer of DNA in the centrifugation tube. After one replication, there is only one layer (mix of 14N and of 15N). After two replication, there are two layers in the tube (14-15N and 14N). It confirms that the replication is semi-conservative.
The replication of the DNA is done by the DNApolymerase, which takes one strand of DNA as model and replicates it from 5’ à 3’, adding one nucleotide by one. This addition is done from deoxynucleosides triphosphate and generate an inorganic phosphate (or pyrophosphate)
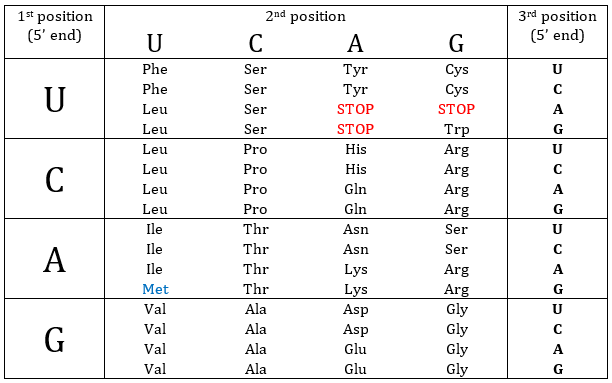
Nucleotides form triplets, called codons. The table below shows the codons and the names we call them with.
Determination of the sequence: the method of Sanger
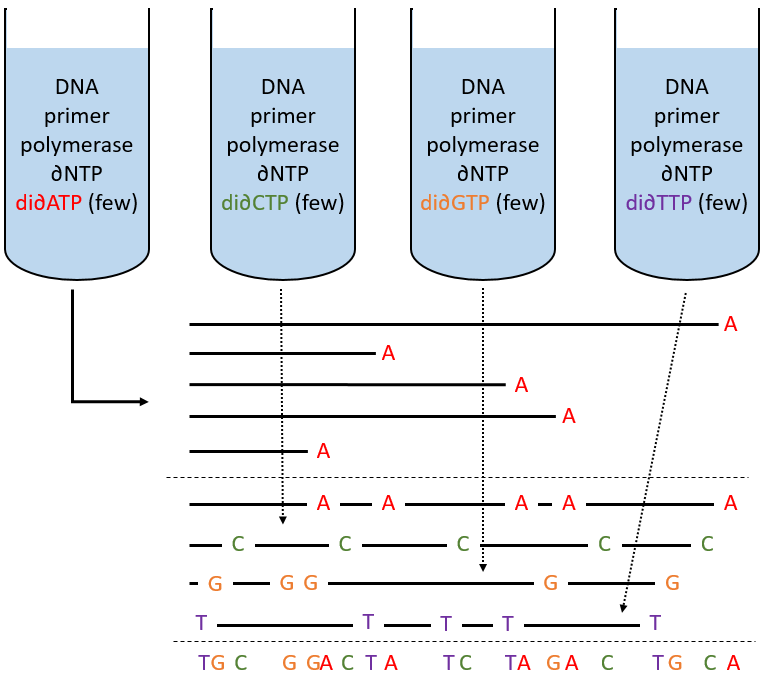
To use this method, we need a complete strand of DNA and one fragment of it obtained via an endonuclease. The fragment is called a primer. We also need to prepare 4 different solutions. In each there are some DNA polymerases and some of each ∂NTP. One of the ∂NTP is marked. In the first solution, we add a few di∂ATP. In this dideoxynucleotide, the OH of the 3’ is missing and it stops the chain. In the second solution, we place a few di∂CTP, in the third a few di∂GTP and in the fourth a few di∂TTP. When we put the primer and the DNA in one solution, the DNApolymerase synthesises a strand at the extremity of the primer, copying the sequence of the DNA. However the synthesis will stop when one di∂NTP is added to the sequence. In each solution we know which nucleotide ends the sequence but the chains can differ in length: for instance in the first solution if one adenosine was to be added by the DNApolymerase, it can come from a ∂ATP or from a di∂ATP. In the first case the chain continues to grow and not in the second case.
The same happens in the other solutions except that the strand ends by a different nucleotide. We analyse the 4 solutions in parallel on an electrophoresis gel. On this gel, the smallest molecules move faster than the long ones and they can be recognised via to the marked nucleotide. We can thus determine the size of each fragment knowing its final extremity. As a result, the complete sequence can eventually be determined.